Abstract
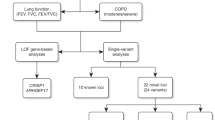
Chronic obstructive pulmonary disease (COPD) is a leading cause of mortality worldwide1. We performed a genetic association study in 15,256 cases and 47,936 controls, with replication of select top results (P < 5 × 10−6) in 9,498 cases and 9,748 controls. In the combined meta-analysis, we identified 22 loci associated at genome-wide significance, including 13 new associations with COPD. Nine of these 13 loci have been associated with lung function in general population samples2,3,4,5,6,7, while 4 (EEFSEC, DSP, MTCL1, and SFTPD) are new. We noted two loci shared with pulmonary fibrosis8,9 (FAM13A and DSP) but that had opposite risk alleles for COPD. None of our loci overlapped with genome-wide associations for asthma, although one locus has been implicated in joint susceptibility to asthma and obesity10. We also identified genetic correlation between COPD and asthma. Our findings highlight new loci associated with COPD, demonstrate the importance of specific loci associated with lung function to COPD, and identify potential regions of genetic overlap between COPD and other respiratory diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Vestbo, J. et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am. J. Respir. Crit. Care Med. 187, 347–365 (2013).
Hancock, D.B. et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nat. Genet. 42, 45–52 (2010).
Repapi, E. et al. Genome-wide association study identifies five loci associated with lung function. Nat. Genet. 42, 36–44 (2010).
Soler Artigas, M. et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat. Genet. 43, 1082–1090 (2011).
Hancock, D.B. et al. Genome-wide joint meta-analysis of SNP and SNP-by-smoking interaction identifies novel loci for pulmonary function. PLoS Genet. 8, e1003098 (2012).
Soler Artigas, M. et al. Sixteen new lung function signals identified through 1000 Genomes Project reference panel imputation. Nat. Commun. 6, 8658 (2015).
Wain, L.V. et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. Lancet Respir. Med. 3, 769–781 (2015).
Fingerlin, T.E. et al. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat. Genet. 45, 613–620 (2013).
Fingerlin, T.E. et al. Genome-wide imputation study identifies novel HLA locus for pulmonary fibrosis and potential role for auto-immunity in fibrotic idiopathic interstitial pneumonia. BMC Genet. 17, 74 (2016).
González, J.R. et al. A common 16p11.2 inversion underlies the joint susceptibility to asthma and obesity. Am. J. Hum. Genet. 94, 361–372 (2014).
Laurell, C.-B. & Eriksson, S. The electrophoretic α-1-globulin pattern of serum in α-1-antitrypsin deficiency. Scand. J. Clin. Lab. Invest. 15, 132–140 (1963).
Silverman, E.K. et al. Genome-wide linkage analysis of severe, early-onset chronic obstructive pulmonary disease: airflow obstruction and chronic bronchitis phenotypes. Hum. Mol. Genet. 11, 623–632 (2002).
Cho, M.H. et al. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir. Med. 2, 214–225 (2014).
Silverman, E.K. et al. Opportunities and challenges in the genetics of COPD 2010: an International COPD Genetics Conference report. COPD 8, 121–135 (2011).
Mannino, D.M. & Buist, A.S. Global burden of COPD: risk factors, prevalence, and future trends. Lancet 370, 765–773 (2007).
Pillai, S.G. et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 5, e1000421 (2009).
Cho, M.H. et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nat. Genet. 42, 200–202 (2010).
Cho, M.H. et al. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Hum. Mol. Genet. 21, 947–957 (2012).
Hobbs, B.D. et al. Exome array analysis identifies a common variant in IL27 associated with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 194, 48–57 (2016).
Wilk, J.B. et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 5, e1000429 (2009).
Wilk, J.B. et al. Genome-wide association studies identify CHRNA5/3 and HTR4 in the development of airflow obstruction. Am. J. Respir. Crit. Care Med. 186, 622–632 (2012).
Hao, K. et al. Lung eQTLs to help reveal the molecular underpinnings of asthma. PLoS Genet. 8, e1003029 (2012).
Giambartolomei, C. et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 10, e1004383 (2014).
Vasioukhin, V., Bowers, E., Bauer, C., Degenstein, L. & Fuchs, E. Desmoplakin is essential in epidermal sheet formation. Nat. Cell Biol. 3, 1076–1085 (2001).
Sato, Y. et al. The novel PAR-1-binding protein MTCL1 has crucial roles in organizing microtubules in polarizing epithelial cells. J. Cell Sci. 126, 4671–4683 (2013).
Sato, Y. et al. MTCL1 crosslinks and stabilizes non-centrosomal microtubules on the Golgi membrane. Nat. Commun. 5, 5266 (2014).
Uhlén, M. et al. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Ward, L.D. & Kellis, M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 40, D930–D934 (2012).
Ward, L.D. & Kellis, M. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 44, D877–D881 (2016).
Lei, Y. et al. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity 36, 933–946 (2012).
Lei, Y., Wen, H. & Ting, J.P. The NLR protein, NLRX1, and its partner, TUFM, reduce type I interferon, and enhance autophagy. Autophagy 9, 432–433 2013).
Kang, M.J. et al. Suppression of NLRX1 in chronic obstructive pulmonary disease. J. Clin. Invest. 125, 2458–2462 (2015).
Wert, S.E. et al. Increased metalloproteinase activity, oxidant production, and emphysema in surfactant protein D gene-inactivated mice. Proc. Natl. Acad. Sci. USA 97, 5972–5977 (2000).
Lomas, D.A. et al. Serum surfactant protein D is steroid sensitive and associated with exacerbations of COPD. Eur. Respir. J. 34, 95–102 (2009).
Foreman, M.G. et al. Polymorphisms in surfactant protein-D are associated with chronic obstructive pulmonary disease. Am. J. Respir. Cell Mol. Biol. 44, 316–322 (2011).
Mathai, S.K. et al. Desmoplakin variants are associated with idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 193, 1151–1160 (2016).
Pickrell, J.K. et al. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 48, 709–717 (2016).
Washko, G.R. et al. Lung volumes and emphysema in smokers with interstitial lung abnormalities. N. Engl. J. Med. 364, 897–906 (2011).
Chilosi, M., Poletti, V. & Rossi, A. The pathogenesis of COPD and IPF: distinct horns of the same devil? Respir. Res. 13, 3 (2012).
Stanley, S.E. et al. Telomerase mutations in smokers with severe emphysema. J. Clin. Invest. 125, 563–570 (2015).
Soriano, J.B. et al. The proportional Venn diagram of obstructive lung disease: two approximations from the United States and the United Kingdom. Chest 124, 474–481 (2003).
Welter, D. et al. The NHGRI GWAS Catalog, a curated resource of SNP–trait associations. Nucleic Acids Res. 42, D1001–D1006 (2014).
Moffatt, M.F. et al. A large-scale, consortium-based genomewide association study of asthma. N. Engl. J. Med. 363, 1211–1221 (2010).
Thorgeirsson, T.E. et al. Sequence variants at CHRNB3–CHRNA6 and CYP2A6 affect smoking behavior. Nat. Genet. 42, 448–453 (2010).
Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat. Genet. 42, 441–447 (2010).
Finucane, H.K. et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 47, 1228–1235 (2015).
Bulik-Sullivan, B. et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015).
Stanley, S.E. et al. Loss-of-function mutations in the RNA biogenesis factor NAF1 predispose to pulmonary fibrosis–emphysema. Sci. Transl. Med. 8, 351ra107 (2016).
Coram, M.A. et al. Leveraging multi-ethnic evidence for mapping complex traits in minority populations: an empirical Bayes approach. Am. J. Hum. Genet. 96, 740–752 (2015).
Deloukas, P. et al. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat. Genet. 45, 25–33 (2013).
Wood, A.R. et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46, 1173–1186 (2014).
Locke, A.E. et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 518, 197–206 (2015).
Zheng, H.F. et al. Whole-genome sequencing identifies EN1 as a determinant of bone density and fracture. Nature 526, 112–117 (2015).
Castaldi, P.J. et al. The association of genome-wide significant spirometric loci with chronic obstructive pulmonary disease susceptibility. Am. J. Respir. Cell Mol. Biol. 45, 1147–1153 (2011).
Winkler, T.W. et al. Quality control and conduct of genome-wide association meta-analyses. Nat. Protoc. 9, 1192–1212 (2014).
Willer, C.J., Li, Y. & Abecasis, G.R. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Yang, J., Lee, S.H., Goddard, M.E. & Visscher, P.M. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88, 76–82 (2011).
Yang, J. et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 44, 369–375 (2012).
Lamontagne, M. et al. Refining susceptibility loci of chronic obstructive pulmonary disease with lung eqtls. PLoS One 8, e70220 (2013).
Bossé, Y. et al. Molecular signature of smoking in human lung tissues. Cancer Res. 72, 3753–3763 (2012).
Lamontagne, M. et al. Genetic regulation of gene expression in the lung identifies CST3 and CD22 as potential causal genes for airflow obstruction. Thorax 69, 997–1004 (2014).
Langmead, B., Trapnell, C., Pop, M. & Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Chang, C.C. et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2016).
Hardin, M. et al. The clinical and genetic features of COPD–asthma overlap syndrome. Eur. Respir. J. 44, 341–350 (2014).
Wakefield, J. Bayes factors for genome-wide association studies: comparison with P-values. Genet. Epidemiol. 33, 79–86 (2009).
Morris, A.P. Transethnic meta-analysis of genomewide association studies. Genet. Epidemiol. 35, 809–822 (2011).
Kichaev, G. & Pasaniuc, B. Leveraging functional-annotation data in trans-ethnic fine-mapping studies. Am. J. Hum. Genet. 97, 260–271 (2015).
Lu, Q., Powles, R.L., Wang, Q., He, B.J. & Zhao, H. Integrative tissue-specific functional annotations in the human genome provide novel insights on many complex traits and improve signal prioritization in genome wide association studies. PLoS Genet. 12, e1005947 (2016).
Trynka, G. et al. Disentangling the effects of colocalizing genomic annotations to functionally prioritize non-coding variants within complex-trait loci. Am. J. Hum. Genet. 97, 139–152 (2015).
Slowikowski, K., Hu, X. & Raychaudhuri, S. SNPsea: an algorithm to identify cell types, tissues and pathways affected by risk loci. Bioinformatics 30, 2496–2497 (2014).
Tasş an, M. et al. Selecting causal genes from genome-wide association studies via functionally coherent subnetworks. Nat. Methods 12, 154–159 (2015).
International HapMap Consortium. The International HapMap Project. Nature 426, 789–796 (2003).
Cho, M.H. et al. A genome-wide association study of emphysema and airway quantitative imaging phenotypes. Am. J. Respir. Crit. Care Med. 192, 559–569 (2015).
Zhang, K., Cui, S., Chang, S., Zhang, L. & Wang, J. i-GSEA4GWAS: a web server for identification of pathways/gene sets associated with traits by applying an improved gene set enrichment analysis to genome-wide association study. Nucleic Acids Res. 38, W90–W95 (2010).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30 (2000).
Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000).
Gene Ontology Consortium. Gene Ontology Consortium: going forward. Nucleic Acids Res. 43, D1049–D1056 (2015).
Acknowledgements
Please refer to the Supplementary Note for full acknowledgments.
Author information
Authors and Affiliations
Consortia
Contributions
B.D.H. and M.H.C. contributed to the study concept and design, data analysis, statistical support, and manuscript writing. K.d.J., A.B.W., S.J.L., and D.P.S. contributed to the study concept and design and to data analysis. N.S. and M.S.A. contributed to the data analysis and statistical support. T.H.B. and J.E.H. contributed to the study concept and design and to statistical support. L.L. contributed to the data collection, data analysis, and statistical support. K.E.N. contributed to data collection and data analysis. J.D.C., B.M.P., N.L., R.T.-S., G.T.O., Y.T., R.G.B., S.I.R., P.B., A.G., P.G.W., D.A.M., D.A.S., and E.K.S. contributed to the study concept and design and to data collection. D.Q., T.A.F., M.L., Y.B., N.S., N.F., P.J.C., R.P.C., T.M.B., S.A.G., J.C.L., J.D., J.B.W., M.K.L., S.L., A.M., X.-Q.W., and E.J.A. contributed to the data analysis. L.V.W., I.P.H., P.D.P., D.S.P., W.M., M.D.T., and H.M.B. contributed to the study concept and design. S.R.H., M.O., J.V., P.A.D., W.J.K., Y.-M.O., S.S.R., D.S., A.A.L., G.G.B., B.H.S., A.G.U., E.R.B., D.A.L., J.-J.Y., D.K.K., I.H., P.S., and M.H. contributed to the data collection. All authors, including those whose initials are not listed above, contributed to the critical review and editing of the manuscript and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
I.P.H. has received grant support from Pfizer. P.J.C. has received research funding from GlaxoSmithKline. B.P. serves on the data and safety monitoring board (DSMB) of a clinical trial funded by the manufacturer and on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. N.L. and R.T.-S. are shareholders and employees of GlaxoSmithKline. S.I.R. is a current employee and shareholder at AstraZeneca. He has served as a consultant, participated on advisory boards, and received honoraria for speaking or grant support from the American Board of Internal Medicine, Advantage Healthcare, Almirall, the American Thoracic Society, AstraZeneca, Baxter, Boehringer Ingelheim, Chiesi, ClearView Healthcare, the Cleveland Clinic, Complete Medical Group, CSL, Dailchi Sankyo, Decision Resources, Forest, Gerson Lehman, Grifols, GroupH, Guidepoint Global, Haymarket, Huron Consulting, Inthought, Johnson & Johnson, Methodist Health System–Dallas, NCI Consulting, Novartis, Pearl, Penn Technology, Pfizer, PlanningShop, PSL FirstWord, Qwessential, Takeda, Theron, and WebMD. W.T. reports fees to the Department, all outside the submitted work, from Pfizer, GSK, Chiesi, Roche Diagnostics/Ventana, Biotest, Merck Sharp Dohme, Novartis, Lilly Oncology, Boehringer Ingelheim, and grants from Dutch Asthma Fund. J.C.L. is currently an employee of GNS Healthcare. J.B.W. was employed by Pfizer during the time this research was performed. P.B. has received consulting and lecture fees from AstraZeneca, Boehringer Ingelheim, Chiesi, Novartis, and Teva. L.L. has performed consultancy for Boehringer Ingelheim and has received an AstraZeneca Scientific Award and travel support from Novartis, the European Respiratory Society, and the Belgian Respiratory Society. P.G.W. has consulted for Amgen, Sanofi, Novartis, Genentech/Roche, Boehringer Ingelheim, and Neostem and has had research grants from Pfizer and Genentech. D.L. received grant support, honoraria, and consultancy fees from GlaxoSmithKline for work on the ICGN and ECLIPSE studies and was a member of and chaired the GSK Respiratory Therapy Area Board (2009–2015). M.H. is a current employee of AstraZeneca. D.A.S. is serving on the scientific advisory boards of Apellis Pharmaceuticals and Pliant Therapeutics, and is the founder and owner of Eleven P15. The University of Groningen has received money for D.S.P. with regard to a grant for research from AstraZeneca, Chiesi, Genentec, GlaxoSmithKline, and Roche. Fees for consultancies were given to the University of Groningen by AstraZeneca, Boehringer Ingelheim, Chiesi, GlaxoSmithKline, Takeda, and TEVA. E.K.S. has received honoraria and consulting fees from Merck, grant support and consulting fees from GlaxoSmithKline, and honoraria and travel support from Novartis. M.H.C. has received grant support from GlaxoSmithKline.
Additional information
A list of members and affiliations appears in the Supplementary Note.
A list of members and affiliations appears in the Supplementary Note.
A list of members and affiliations appears in the Supplementary Note.
A list of members and affiliations appears in the Supplementary Note.
A list of members and affiliations appears in the Supplementary Note.
A list of members and affiliations appears in the Supplementary Note.
A list of members and affiliations appears in the Supplementary Note.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6, Supplementary Tables 1, 2, 4–7, 9–13 and 15–20, and Supplementary Note (PDF 8454 kb)
Supplementary Table 3
Full results table for all 79 variants submitted for testing in UK BiLEVE stage 2 analysis. (XLSX 63 kb)
Supplementary Table 8
Full results (P < 0.05 in meta-analysis) for lung expression quantitative trait locus (eQTL) analysis. (XLSX 24 kb)
Supplementary Table 14
Lookup of NHGRI-EBI GWAS Catalog asthma-associated trait genome-wide significant GWAS loci in our COPD association stage 1 meta-analysis results. (XLSX 13 kb)
Rights and permissions
About this article
Cite this article
Hobbs, B., de Jong, K., Lamontagne, M. et al. Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nat Genet 49, 426–432 (2017). https://doi.org/10.1038/ng.3752
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng.3752
This article is cited by
-
Associations between long-term night shift work and incidence of chronic obstructive pulmonary disease: a prospective cohort study of 277,059 UK Biobank participants
BMC Medicine (2024)
-
Human genetic associations of the airway microbiome in chronic obstructive pulmonary disease
Respiratory Research (2024)
-
Genetics of chronic respiratory disease
Nature Reviews Genetics (2024)
-
Deep learning model improves COPD risk prediction and gene discovery
Nature Genetics (2023)
-
A genome-wide association study identifies distinct variants associated with pulmonary function among European and African ancestries from the UK Biobank
Communications Biology (2023)