Abstract
Inflammasomes are multiprotein complexes that activate caspase-1, which leads to maturation of the proinflammatory cytokines interleukin 1β (IL-1β) and IL-18 and the induction of pyroptosis. Members of the Nod-like receptor (NLR) family, including NLRP1, NLRP3 and NLRC4, and the cytosolic receptor AIM2 are critical components of inflammasomes and link microbial and endogenous danger signals to the activation of caspase-1. In response to microbial infection, activation of the inflammasomes contributes to host protection by inducing immune responses that limit microbial invasion, but deregulated activation of inflammasomes is associated with autoinflammatory syndromes and other pathologies. Thus, understanding inflammasome pathways may provide insight into the mechanisms of host defense against microbes and the development of inflammatory disorders.
Similar content being viewed by others
Main
The innate immune system is the first line of defense against microbial infection and is activated by the engagement of germline-encoded pattern-recognition receptors (PRRs) in response to microbes1. PRRs recognize the presence of unique microbial components called pathogen-associated molecular patterns or endogenous damage-associated molecular patterns generated in the setting of cellular injury or tissue damage2. In response to infection, PRR activation initiates signal-transduction pathways that ultimately culminate in host defense responses that eliminate microbial invasion. A major inflammatory pathway is activation of the inflammasome, a multiprotein platform that activates caspase-1 (ref. 3). Once activated, caspase-1 proteolytically cleaves the cytokine precursors pro-interleukin 1β (pro-IL-1β) and pro-IL-18; this is critical for release of the biologically active forms (L-1β and IL-18) and triggers proinflammatory and antimicrobial responses4. Active caspase-1 can also cleave less-defined protein substrates to regulate the induction of pyroptosis, autophagy and bacterial degradation through mechanisms that are poorly understood4. So far, four inflammasomes have been identified; these are named after the PRR that regulates their activity: NLRP1, NLRP3, NLRC4 and AIM2. Except for AIM2, these inflammasomes contain a PRR that belongs to the Nod-like receptor (NLR) family. NLRs are intracellular PRRs defined by a tripartite structure comprising the following: anamino-terminal caspase-recruitment domain (CARD), pyrin domain, acidic transactivating domain or baculovirus inhibitor repeat that mediates downstream protein-protein interactions; a central nucleotide-binding-and-oligomerization domain (Nod) that mediates self-oligomerization; and carboxy-terminal leucine-rich repeats (LRRs) that are thought to sense different microbial and endogenous damage stimuli4. In this review, we focus on the activation, regulation and function of NLR inflammasomes with an emphasis on their interaction with microbes and their role in host defense.
NLRC4 inflammasome: microbe recognition and activation
NLRC4 is important for the activation of caspase-1 in macrophages infected with pathogenic bacteria, including Salmonella enterica serovar Typhimurium (S. enterica)5,6, Legionella pneumophila7,8,9 and Pseudomonas aeruginosa10,11. The activation of caspase-1 by these pathogenic bacteria requires a functional bacterial secretion system that has been suggested as a link between bacterial pathogenicity and NLRC4 activation3. These secretion systems, which include the type III secretion system (T3SS) and type IV secretion system (T4SS), act as molecular needle-like structures that inject effector proteins into the cytosol of host cells and are critical for pathogen colonization. Flagellin, the main component of the flagellum, is important for activation of the NLRC4 inflammasome5,6. Because the delivery of purified flagellin to the macrophage cytosol triggers caspase-1 activation through NLRC4 (refs. 5,6), it had been thought that NLRC4 is activated in macrophages via the leakage of small amounts of flagellin through a T3SS (for example, S. enterica and P. aeruginosa) or T4SS (for example, L. pneumophila) during infection12. However, Shigella flexneri, an aflagellated pathogenic bacterium, also induces activation of the NLRC4 inflammasome through the T3SS13. Furthermore, flagellin-deficient S. enterica and P. aeruginosa can activate NLRC4 at high ratios of bacteria to macrophages, which further suggests that factors other than flagellin can induce activation of the NLRC4 inflammasome14,15. Initial insights into the flagellin-independent pathway were provided by the observation that proteins that form the basal body rod component of the T3SS, such as PrgJ, can activate the NLRC4 inflammasome. PrgJ-like proteins contain regions structurally homologous to the carboxy-terminal portion of flagellin14, which is a critical portion of flagellin that is sufficient to trigger NLRC4 inflammasome activation6,16. However, the contribution of T3SS rod proteins to activation of the NLRC4 inflammasome is difficult to evaluate because bacterial pathogens without a functional secretion system are impaired in the secretion of effector proteins and are therefore highly attenuated.
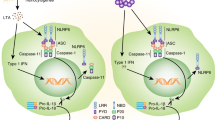
How does NLRC4 sense different structures such as flagellin and PrgJ-like proteins? Initial studies have shown a link between NLRC4 and Naip5 (neuronal apoptosis inhibitor protein 5), another member of the NLR family. Naip5 senses the carboxy-terminal region of flagellin and is required for the activation of NLRC4 in response to L. pneumophila16. In contrast, Naip5 is dispensable for NLRC4 activation in response to S. enterica, P. aeruginosa or flagellin purified from S. enterica16. Those puzzling results have been clarified by the observation that distinct Naip proteins link flagellin and PrgJ-like proteins to NLRC4. Whereas flagellin binds Naip5 and Naip6, PrgJ-like proteins interact with Naip2 (refs. 17,18). CprI, a subunit of the secretion system of Chromobacterium violaceum, binds to human NAIP17. Results obtained by reconstitution of the NLRC4 inflammasome in 293 human embryonic kidney cells also suggest that Naip proteins act upstream of NLRC4 to promote inflammasome activation17,18. Although the mechanism by which Naip proteins activate NLRC4 remains unclear, one model proposes that flagellin or PrgJ-like proteins bind to the LRRs of Naip proteins to induce a conformational change in the latter; this in turn induces the activation of NLRC4 (Fig. 1). A better understanding of the link between Naip proteins and NLRC4 is needed to explain why Naip5 is essential for the activation of caspase-1 in response to L. pneumophila but is dispensable in S. enterica infection despite the fact that flagellin from both pathogens binds to Naip5 (refs. 16,17,18). Because the amino terminus of flagellin can inhibit the interaction of flagellin with Naip5, the differences in the role of Naip5 in the recognition of L. pneumophila and S. enterica could depend on other factors that regulate the exposure or conformation of the carboxyl terminus, the portion of flagellin that is sufficient for NLRC4 activation19. Finally, S. enterica potently induces caspase-1 activation in human cells20, but flagellin is not sensed by human NAIP17, which raises questions about the molecular mechanism that induces caspase-1 activation in human cells.
Infection of macrophages with various Gram-negative bacteria, including S. enterica, L. pneumophila and P. aeruginosa, activates caspase-1 via NLRC4. A critical step is the cytosolic delivery of flagellin or PrgJ-like proteins through bacterial T3SS or T4SS. Flagellin is recognized by Naip5 or Naip6 (not shown here), whereas PrgJ-like proteins are recognized by Naip2. S. flexneri activates the NLRC4 inflammasome independently of flagellin through an unknown microbial product. Activation of caspase-1 via NLRC4 leads to the processing and release of IL-1β and IL-18, the processing of caspase-7 and the induction of other cellular activities that are poorly understood.
Role of the NLRC4 inflammasome in host defense
The NLRC4 inflammasome regulates host defense by controlling the release of IL-1β and IL-18, bacterial degradation and pyroptosis. IL-1β and IL-18 have an important role in the host defense response to S. flexneri21, but it is unknown whether the production of these cytokines depends on NLRC4 in vivo. In the case of S. enterica infection, caspase-1 and IL-18 both have a role in host defense22,23. NLRC4 does not have a role in host defense after orogastric infection in C57BL/6 mice23,24, but it confers host protection to mice on the BALB/c background (lethal factor; G.N., unpublished observations). It was initially observed that activators of the NLRC4 inflammasome induce pyroptosis, a form of caspase-1-dependent cell death with features of both apoptosis and necrosis25. Notably, S. enterica represses expression of flagellin and T3SS during the systemic phase of infection, and downregulation of these factors prevents activation of the NLRC4 inflammasome26. In fact, mutants of S. enterica26 or L. monocytogenes27,28 that have enforced expression of flagellin cannot evade detection by the NLRC4 inflammasome and are highly attenuated. Furthermore, NLRC4 restricts S. enterica strains that overexpress flagellin independently of IL-1β and IL-18 by promoting the release of intracellular S. enterica from pyroptotic macrophages; these bacteria are consequently engulfed and killed by neighboring neutrophils26. However, the contribution of this protective mechanism to host defense during physiological infection with wild-type intracellular bacteria remains to be determined.
NLRC4 can also promote the degradation of pathogens inside macrophages. This is exemplified by studies of L. pneumophila, a Gram-negative intracellular bacterium that causes Legionnaires' disease, an acute form of pneumonia. After infection, L. pneumophila replicates in specialized vacuoles inside macrophages. Notably, NLRC4-dependent activation of caspase-1 restricts the intracellular growth of L. pneumophila at least in part by promoting the fusion of bacteria-containing vacuoles with lysosomes9. The ability to restrict L. pneumophila growth by NLRC4 requires expression of flagellin and host caspase-7, a proteolytic substrate of caspase-1, but not IL-1β and IL-18 (refs. 9,29). However, the mechanism by which caspase-7 promotes the fusion of L. pneumophila–containing vacuoles with lysosomes and bacterial degradation is unclear. Naip5 also has a critical role in restricting the growth of L. pneumophila inside macrophages; this can be partially explained by a role of Naip5 in linking cytosolic flagellin to NLRC4 activation9,29,30. However, analysis of A/J mice that express a mutant Naip5 protein that supports the replication of L. pneumophila and Naip5-null mice suggests that Naip5 may also act independently of NLRC4 to regulate L. pneumophila replication31. Although both mutant mice show greater bacterial replication than wild type do, the amino acids of Naip5 substituted in A/J mice are not important for flagellin binding or activation of the NLRC4 inflammasome17, which suggests that Naip5 has a role in restricting bacterial replication that is distinct from its role in activating the NLRC4 inflammasome. In addition to promoting bacterial degradation, NLRC4 and Naip5 can control L. pneumophila growth through the induction of pyroptosis, a phenotype evident under high multiplicity of infection9,30. Whereas macrophages deficient in Naip5, NLRC4, caspase-1 or caspase-7 show a notable phenotype in regulating L. pneumophila replication in vitro, the role of the flagellin-Naip5-NLRC4 signaling pathway in lung infection is more modest, whichsuggests that other signaling pathways can compensate for inflammasome deficiency in vivo9,29,32.
The NLRP1 inflammasome
The initial description of the inflammasome was based on the assembly of the human NLRP1 inflammasome. Although the role of NLRP1 in immune responses remains poorly understood, its relevance is underscored by the association of variations of the gene encoding NLRP1 with generalized vitiligo, vitiligo-associated type I diabetes, Addison's disease, rheumatoid arthritis and Alzheimer's disease3. The domain structure of human NLRP1 comprises an amino-terminal pyrin domain, a centrally located Nod and LRRs, and carboxy-terminal FIINDs (function-to-find domains) and CARDs. Thus, NLRP1 differs from other NLRs proteins by having two signal-transduction domains; that is, a pyrin domain and CARD. Analysis of extracts of the THP-1 human myeloid cell line with macrophage-like properties has shown that NLRP1 forms a multiprotein complex containing the adaptor ASC, CARD8, caspase-5 and caspase-1 that has IL-1β-processing activity33. Subsequently, reconstitution of the NLRP1 inflammasome with purified components has shown that the minimal elements of the NLRP1 inflammasome are NLRP1, an NTP and caspase-1 (ref. 34). The activity of the reconstituted NLRP1 inflammasome is induced by muramyl dipeptide (MDP), and on the basis of those initial studies, caspase-1 has been proposed to be activated through a two-step mechanism: first, microbial MDP binds NLRP1 and changes its conformation, which allows it to bind an NTP; this in turn induces oligomerization of NLRP1 through its Nod, thus creating a platform for caspase-1 activation. The adaptor ASC is not essential for caspase-1 activation, probably because human NLRP1 can bind to caspase-1 directly through a CARD-CARD interaction, but the presence of ASC augments NLRP1-mediated caspase-1 activation34. Consistent with those results, macrophages stimulated with MDP in complex with titanium dioxide activate the NLRP1 inflammasome in an ASC-independent manner35. However, there is no direct evidence that MDP binds to NLRP1. Notably, NLRP1 and CARD8 are proteolytically processed, but it is still unclear whether these events are necessary for caspase-1 activation36. Thus, further work is needed to understand the activation of NLRP1 and the role of CARD8 and caspase-5 in the function of the NLRP1 inflammasome.
Unlike humans, who have a single NLRP1 gene, mice have three tandem paralogs, Nlrp1a, N1rp1b and Nlrp1c-ps (also known as Nalp1a, Nalp1b and Nalp1c, respectively). Furthermore, different strain-specific alleles exist for Nlrp1b, and these genetic variants have led to the identification of NLRP1b as the sensor of Bacillus anthracis lethal toxin37. Lethal toxin, a metalloproteinase, is a bipartite toxin that consists of protective antigen and a pore-forming molecule that mediates the translocation of lethal factor into the host cytosol, where it activates caspase-1. Lethal toxin–induced caspase-1 activation, IL-1β production and pyroptosis require the susceptible Nlrp1b allele37, whereas ASC is required for lethal toxin–induced production of IL-1β but is dispensable for pyroptosis38. In vivo experiments have shown that the product of Nlrp1b allelic variation has a protective role in B. anthracis infection and, consistent with a role for NLRP1b in activating the inflammasome, mice deficient in caspase-1 (Casp1−/−) or IL-1β (Il1b−/−) are more susceptible to B. anthracis infection39. Thus, the host-protective response mediated via the NLRP1B inflammasome depends on the production of IL-1β rather than the induction of pyroptosis. Although much progress has been made in elucidating the recognition of B. anthracis via the NLRP1B inflammasome, the mechanism by which lethal factor triggers activation of the NLRP1B inflammasome remains elusive.
The NLRP3 inflammasome: mechanism of activation
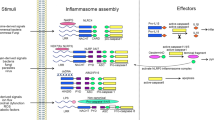
Caspase-1 was initially identified as the protease responsible for the maturation and release of IL-1β in response to ATP. Deeper understanding of this process was provided by the discovery that NLRP3 and ASC are required for the activation of caspase-1 in response to ATP and certain bacterial pore-forming toxins3. Consistent with the importance of NLRP3 in IL-1β production, NLRP3 gain-of-function mutations lead to cryopyrin-associated periodic syndromes, which are efficiently treated with inhibitors of IL-1β-mediated signaling3. The NLRP3 inflammasome is activated by a plethora of microbial stimuli, including MDP40,41, bacterial RNA42, the double-stranded RNA analog poly(I:C)42, lipopolysaccharide, microbial lipopeptide, the imiquimod R-837 and the synthetic imidazoquinoline resiquimod (R-848)42. NLRP3 can also be activated by endogenous stimuli and particulate matter, such as uric acid, cholesterol and hydroxyapatite crystals, silica, aluminum salts, asbestos, malarial hemozoin, amyloid deposits and fatty acids4. Given the chemical and structural diversity of the NLRP3 activators, it has been hypothesized that NLRP3 does not interact directly with its activators; instead, its activation is triggered through an intermediate cellular signal elicited by all these stimuli. Evidence indicates that most, if not all, Toll-like receptor (TLR) agonists and MDP do not directly activate the NLRP3 inflammasome. Instead, these microbial stimuli prime the activation of NLRP3 through the induction of NLRP3 expression in macrophages43, a prerequisite for inflammasome activation43,44,45. Consistent with that, activation of the NLRP3 inflammasome by ATP, bacterial pore-forming toxins and particulate matter requires prestimulation with TLR agonists to induce NLRP3 expression43,44,45,46. Whereas the adaptor TRIF has a minor role in the priming process in response to stimulation with lipopolysaccharide47, it has a major role in response to bacterial RNA48. Because NLRP3 induction is mediated by the transcription factor NF-κB, endogenous cytokines such as tumor-necrosis factor and IL-1β are also effective in inducing NLRP3 expression and promoting caspase-1 activation in response to NLRP3 activators45. Thus, activation of the NLRP3 inflammasome requires two signals in mouse macrophages. The first signal is provided by microbial or endogenous molecules that activate NF-κB and induce NLRP3 expression (Fig. 2). The second signal directly activates NLRP3 and is provided by ATP, certain bacterial toxins or particulate matter (Fig. 2). The situation may be different in human monocytes and microglia cells, in which stimulation with TLR ligands induces the release of IL-1β without exogenous stimulation with ATP49,50,51. However, stimulation of human monocytes and microglia cells with TLR has been proposed to induce the release of endogenous ATP that acts in an autocrine way to activate the ion channel P2X7 (refs. 49,50,51). Notably, under conditions in which the autophagic pathway is compromised, stimulation with pathogen-associated molecular patterns induces activation of the NLRP3 inflammasome52 and more production of pro-IL-1β53,54, which suggests that autophagy has a role in controlling the production of IL-1β.
Activation of caspase-1 via NLRP3 requires two signals. Signal 1 is represented by microbial molecules or endogenous cytokines and is required for the upregulation of NLRP3 and pro-IL-1β. Signal 2 activates the NLRP3 inflammasome. Activation by S. aureus, S. pyogenes, S. pneumoniae and V. cholerae is mediated by pore-forming toxins. Other bacterial toxins can also induce the activation of the NLRP3 inflammasome, such as cholera toxin (CT) or C. difficile toxins TcdA and TcdB (not shown here); C. albicans induces activation of the NLRP3 inflammasome through the kinase Syk, although the mechanism involved is unclear. Influenza virus can induce the activation of the NLRP3 inflammasome, but it is controversial whether this is due to a pore-forming activity mediated by M protein or to sensing of viral RNA species in the cytosol. Cytosolic bacterial RNA induces activation of the NLRP3 inflammasome. TNFR, receptor for tumor-necrosis factor.
Several theories have been proposed for the identity of the cellular signal responsible for NLRP3 activation, including a change in the intracellular concentration of K+ and Na+, the formation of a large pore in cell membranes, the release of cathepsins from damaged lysosomes, the production of reactive oxygen species (ROS) and damage in the mitochondria3. The involvement of K+ efflux in NLRP3 activation is supported by the fact that some NLRP3 activators, including ATP, the antibiotic nigericin and pore-forming toxins, results in lower intracellular concentration of K+, and a high extracellular concentration of K+ inhibits activation of the NLRP3 inflammasome55,56. However, there is no evidence that particulate matter triggers efflux of K+. Furthermore, the interpretation of experiments in which extracellular Na+ is replaced with K+ is complicated because extracellular Na+ is also reported to be required for NLRP3 activation independently of K+ efflux57. Hence, it is difficult to discern whether the inhibitory effect of isotonic medium with a high concentration of K+ on NLRP3 activation is due to the high concentration of K+ or the low concentration of Na+. Very high extracellular concentrations of K+ also block the activation of the NLRP1, NLRC4 and AIM2 inflammasomes58. Therefore, further studies are required to clarify the role of changes in cytosolic ionic concentrations in the activation of the NLRP3 inflammasome.
Extracellular ATP activates NLRP3 through the opening of ATP-gated P2X7. In contrast, bacterial pore-forming toxins activate NLRP3 independently of P2X7 (refs. 44,59). P2X7 is unique among ion channels in that its activation not only opens a cation channel but also leads to the opening of a larger pore permeable to molecules 900 Da or greater in size. It has been suggested that the opening of a large pore formed by the hemichannel pannexin-1 after P2X7 stimulation is necessary for NLRP3 activation independently of K+ efflux60. However, no defect in NLRP3 activation or opening of the large pore in pannexin-1-deficient macrophages stimulated with ATP and nigericin has been found61. Although those results suggest that pannexin-1 is not the molecular component of the large pore opened by ATP, it is still unknown whether the opening of a large pore is required to activate NLRP3.
The activation of NLRP3 by particulate matter requires endocytosis, as pretreatment of macrophages with drugs that interfere with cytoskeletal dynamics (such as colchicine and cytochalasin B) inhibit the activation of NLRP3 by uric acid crystals, silica and aluminum salts but not by ATP4. Furthermore, inhibitors of cathepsin B can prevent the activation of caspase-1 induced by certain microbes62,63. However, as cathepsin B–deficient mice have a modest or no defect in the activation pf NLRP3 by particulate matter64, the observed impairment of NLRP3 activation by the inhibitor of cathepsin B could be due to off-target effects. Alternatively, given the considerable redundancy among the members of the cathepsin family, several lysosomal proteases may be able to trigger NLRP3 activation. Studies of mice with genetic double deficiency could help clarify the role of cathepsins in NLRP3 activation and the mechanism involved.
The production of ROS has also been suggested to act as a common cellular signal upstream of NLRP3 triggered by ATP and particulate matter65. NLRP3 activation is blocked by ROS scavengers and NAPDH-oxidase inhibitors65. In line with those findings, thioredoxin-interaction protein has been proposed to bind and activate NLRP3 after the production of ROS by NLRP3 activators66. However, those results were not independently confirmed in a different study67. Furthermore, another report has suggested that ROS inhibitors interfere with NLRP3 priming rather than its activation46. ROS derived from the mitochondria have been suggested to mediate activation of the NLRP3 inflammasome in a study using inhibitors of the respiratory chain68. However, studies with chemical inhibitors, especially at high concentrations, are prone to artifacts. The release of mitochondrial DNA induced by activators of the NLRP3 inflammasome further amplifies caspase-1 activation69. Although none of the aforementioned results provides a satisfactory explanation for the mechanism of NLRP3 activation, a combination of several cellular signals could be required for NLRP3 activation. Indeed, NLRP3 has been proposed to integrate signals that indicate cellular damage or stress, including membrane permeation, lysosomal damage, ROS production and mitochondrial damage.
NLRP3 activation by microbes
Many bacterial pathogens activate the NLRP3 inflammasome through the secretion of pore-forming toxins. Staphylococcus aureus α-hemolysin activates the NLRP3 inflammasome in combination with TLR2 stimulation by bacterial lipopeptides released during growth59. Analysis of isogenic single-, double- and triple-mutant S. aureus strains defective in α-, β- and γ-hemolysins has shown that these have a redundant role in NLRP3 activation47,59. In vivo experiments with an S. aureus subcutaneous abscess model have shown a critical role for ASC and IL-1β signaling in neutrophil recruitment and control of the infection70,71. Similar to the redundant role of S. aureus hemolysins in NLRP3 activation, Vibrio cholerae secrete the hemolysins HlyA and MARTX to activate NLRP3 (ref. 72). Furthermore, in vivo studies of mouse strains deficient in inflammasome components have shown that caspase-1 and ASC have a protective role against V. cholerae, but NLRP3 does not72. These results suggest that multiple inflammasomes contribute to host defense against V. cholerae in vivo.
Streptococcus pneumoniae, which colonizes the upper respiratory tract, is a leading cause of pneumonia and meningitis and activates NLRP3 through the secreted pore-forming toxin pneumolysin73,74. In an S. pneumoniae lung-infection model73,74, NLRP3 elicits a protective immune response, as Nlrp3−/− mice have higher bacterial loads and greater mortality than do wild-type mice. In contrast, Nlrp3−/− and ASC-deficient (Pycard−/−) mice develop less brain inflammation and have a better clinical outcome than that of wild-type mice in a pneumococcal meningitis model75. Blockade of caspase-1-mediated signaling with a combined regimen of recombinant IL-1 receptor antagonist and IL-18-binding protein leads to considerable amelioration of disease severity and brain pathology, which suggests that interfering with inflammasome activation might be a strategy for treating pneumococcal meningitis.
Several other bacterial toxins induce activation of the NLRP3 inflammasome, including cholera toxin B, adenylcyclase toxin76 and Clostridium difficile toxins A and B77. As these toxins have different mechanisms of action, it remains unclear how they mediate activation of the NLRP3 inflammasome. Such activation mediated by cholera toxin B depends on caspase-11, but activation mediated by adenylcyclase toxin, C. difficile toxin B or pore-forming toxins does not76. Notably, caspase-11 is dispensable for the activation of caspase-1 by most stimuli that activate the NLRP3 inflammasome but has a role in the induction of pyroptosis and release of damage-associated molecular patterns76. Some T3SS effector proteins induce activation of the NLRP3 inflammasome. For example, activation by the Yersinia pestis subspecies Kim has been described for the effector protein YopJ78, an acetyltransferase that causes apoptosis through inactivation of kinases such as MAPKs and IKKs79. The role of NLRP3 in Y. pestis infection in vivo, however, remains to be determined.
Several studies have reported a role for NLRP3 in the innate immune response to viruses. Initial studies showed that the NLRP3 inflammasome can be activated in vitro by Sendai virus80, influenza A virus80 and adenovirus81. Influenza A virus activates NLRP3 through the proton-selective M2 channel82 and elicits a protective inflammatory response83,84,85. However, there is conflicting evidence about the contribution of NLRP3 to the control of viral burden, host survival and the generation of adaptive immunity after influenza infection83,84,85. In an initial analysis83, mice deficient in ASC, caspase-1 or the IL-1 receptor had more mortality accompanied by lower immunoglobulin responses than wild-type mice had after infection with influenza virus, but those deficient in NLRP3 did not. However, in two subsequent studies84,85, Nlrp3−/−, Pycard−/− and Casp1−/− mice had more mortality than wild-type mice but no defect in the generation of adaptive immunity to influenza84. The reason for these contradictory results is unclear.
Candida albicans, a fungus that can cause severe opportunistic infections in immunocompromised hosts, can activate the NLRP3 inflammasome86. Experiments with C. albicans at various morphological stages87,88 and mutants that cannot form hyphae have shown that the yeast form is a more potent activator of NLRP3 than is the hyphal form; furthermore, the transition from yeast to hyphal form is an important step in eliciting NLRP3 activation87. Responsiveness to C. albicans requires TLR2, the receptor for dectin-1, and the signaling molecule Syk and its downstream adaptor CARD9 for the priming step, whereas Syk is required for activation of the NLRP3 inflammasome but CARD9 is not86,88. In vivo experiments with Tlr2−/−, dectin-1-deficient, Nlrp3−/−, Pycard−/−, Casp1−/− and IL-1 receptor–deficient mice have demonstrated a protective role for the NLRP3 inflammasome in a model of disseminated candidiasis88. Notably, both NLRP3 and NLRC4 are required for host defense in a model of oral candidiasis89. However, the mechanism by which C. albicans activates the NLRC4 inflammasome is unclear.
Malaria is caused by Plasmodium parasites, which feed on erythrocyte hemoglobin and use a heme-detoxification mechanism that leads to the formation of dark-brown crystals called 'hemozoin'. As with other particulate matter, there is evidence that hemozoin crystals activate the NLRP3 inflammasome after phagocytosis64. Whereas one study found a modest but important role for NLRP3 in promoting cerebral malaria90, subsequent studies have found no evidence for contribution of NLRP3, ASC, caspase-1, IL-1β or IL-18 in the development of cerebral malaria91, so the role of the NLRP3 inflammasome in this disease remains controversial.
Redundancy in inflammasome activation
Bacterial infection can trigger the activation of several inflammasomes. The clearest example is infection by the intracellullar pathogen Listeria monocytogenes. Initial studies suggested that L. monocytogenes induces activation of the NLRP3 inflammasome, whereas other studies have found NLRP3 to be dispensable for the activation of caspase-192. That apparent discrepancy has been reconciled by the observation that L. monocytogenes can engage multiple inflammasomes and that the contribution of each inflammasome (NLRP3, NLRC4 or AIM2) depends in part on the experimental conditions of infection93,94,95,96. Although these results are controversial, it has been reported that Casp1−/− and Pycard−/− mice are more susceptible than wild-type mice are to L. monocytogenes infection27,97,98. However, it is unclear whether a particular inflammasome is dominant or whether multiple inflammasomes are redundantly activated in vivo. As with L. monocytogenes, several inflammasomes can be activated, depending on the experimental conditions, by other bacteria, including S. enterica5,6,24 and S. flexneri13,14,99, as well as by fungi89 (Table 1). However, except for S. enterica, for these pathogens, the specific contribution of each inflammasome to host defense in vivo remains largely unknown. Mice deficient in both NLRC4 and NLRP3 (Nlrc4−/−Nlrp3−/− mice) are slightly more susceptible to infection with S. enterica than wild-type mice are, but those deficient in either inflammasome alone are not; this is correlated with a five- to ten-fold greater pathogen burden24. Consistently, the phenotype of Nlrc4−/−Nlrp3−/− mice is similar to that of Casp1−/− mice23,24. The role of ASC in host defense against infection with S. enterica is more complex. ASC comprises a pyrin domain and a CARD and is thought to be an essential adaptor that connects NLRP3 to caspase-1 (ref. 3). Experiments with mice deficient in ASC have shown that ASC is necessary for the activation of caspase-1 and the maturation of IL-1β after infection of mice with S. enterica, P. aeruginosa or L. pneumophila92. Notably, ASC is dispensable for the induction of pyroptosis6,11,38, which in these infection models depends on caspase-1 but not on caspase-11 (ref. 38). Studies have shown that induction of pyroptosis does not require the proteolytic maturation of caspase-1 and suggest that phagocytes can assemble two different inflammasomes in response to infection with S. enterica38. One inflammasome containing NLRC4 and caspase-1 is responsible for the induction of pyroptosis, whereas the other inflammasome, containing NLRC4, ASC and caspase-1, mediates the maturation of IL-1β and IL-18 (ref. 38). These data suggest that microbial infections can activate different NLRC4-containing inflammasomes that exert different functions. More detailed analysis of the composition and biochemical properties of different protein complexes in the NLRC4 inflammasome is needed to determine the relevance of these findings.
Pathogen-specific recognition by the inflammasome
Commensal microorganisms abundant in the skin and intestines continuously challenge the immune system without eliciting an inflammatory response. TLRs detect microbial ligands present in the extracellular environment and are activated by both commensal and pathogenic bacteria. However, the keratinized epithelium of the skin and the mucus layer of the gut form a physical barrier that prevents noninvasive microbes from engaging TLRs. In contrast to TLRs, NLR proteins sense the presence of microbial ligands in the cytosol. Thus, members of this class of PRRs are ideal sensors of pathogenic bacteria because bacterial secretion systems or pore-forming toxins, which are features of pathogenic bacteria, can promote the delivery of microbial ligands to the host cytosol. The presence of bacterial secretion systems and pore-forming toxins is important for activation of the inflammasome and production of IL-1β. For example, activation of the NLRC4 inflammasome by pathogenic bacteria requires a functional T3SS or T4SS92. Similarly, activation of the NLRP3 inflammasome by S. aureus, V. cholerae and Streptococcus pyogenes requires bacterial pore-forming toxins4,92. In contrast to peripheral tissues, the intestine is populated with specialized resident phagocytes that are hyporesponsive to microbial stimulation100. It will be important to determine whether the inflammasomes have a role in the discrimination of pathogenic versus nonpathogenic bacteria in the gut.
Conclusions and future perspectives
In the past decade, much progress has been made in elucidating the activation, regulation and function of the inflammasomes in response to microbes. Researchers have identified the microbial sensors responsible for the activation of caspase-1 and a role for Naip proteins in promoting NLRC4 activation. Moreover, there is conclusive evidence that the inflammasomes contribute to host defense against a variety of pathogens. However, the molecular mechanism by which Naip proteins activate NLRC4 and different stimuli induce activation of the NLRP3 inflammasome remains largely unknown. Furthermore, little is known about the protein substrates cleaved by caspase-1 and/or caspase-11 and their role in executing pyroptosis. Another unresolved question is how inflammasomes and other signaling pathways act together in vivo to orchestrate innate and adaptive immune responses. Clearly, much remains to be learned about the inflammasomes and their role in microbial recognition and host defense against microbes.
References
Kumar, H., Kawai, T. & Akira, S. Pathogen recognition by the innate immune system. Int. Rev. Immunol. 30, 16–34 (2011).
Chen, G.Y. & Nunez, G. Sterile inflammation: sensing and reacting to damage. Nat. Rev. Immunol. 10, 826–837 (2010).
Franchi, L., Eigenbrod, T., Munoz-Planillo, R. & Nunez, G. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat. Immunol. 10, 241–247 (2009).
Franchi, L., Warner, N., Viani, K. & Nunez, G. Function of Nod-like receptors in microbial recognition and host defense. Immunol. Rev. 227, 106–128 (2009).
Miao, E.A. et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat. Immunol. 7, 569–575 (2006).
Franchi, L. et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1β in Salmonella-infected macrophages. Nat. Immunol. 7, 576–582 (2006).
Zamboni, D.S. et al. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat. Immunol. 7, 318–325 (2006).
Ren, T., Zamboni, D.S., Roy, C.R., Dietrich, W.F. & Vance, R.E. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2, e18 (2006).
Amer, A. et al. Regulation of Legionella phagosome maturation and infection through flagellin and host Ipaf. J. Biol. Chem. 281, 35217–35223 (2006).
Miao, E.A., Ernst, R.K., Dors, M., Mao, D.P. & Aderem, A. Pseudomonas aeruginosa activates caspase 1 through Ipaf. Proc. Natl. Acad. Sci. USA 105, 2562–2567 (2008).
Franchi, L. et al. Critical role for Ipaf in Pseudomonas aeruginosa–induced caspase-1 activation. Eur. J. Immunol. 37, 3030–3039 (2007).
Sun, Y.H., Rolan, H.G. & Tsolis, R.M. Injection of flagellin into the host cell cytosol by Salmonella enterica serotype Typhimurium. J. Biol. Chem. 282, 33897–33901 (2007).
Suzuki, T. et al. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 3, e111 (2007).
Miao, E.A. et al. Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc. Natl. Acad. Sci. USA 107, 3076–3080 (2010).
Sutterwala, F.S. et al. Immune recognition of Pseudomonas aeruginosa mediated by the IPAF/NLRC4 inflammasome. J. Exp. Med. 204, 3235–3245 (2007).
Lightfield, K.L. et al. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat. Immunol. 9, 1171–1178 (2008).
Zhao, Y. et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 477, 596–600 (2011).
Kofoed, E.M. & Vance, R.E. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 477, 592–595 (2011).
Lightfield, K.L. et al. Differential requirements for NAIP5 in activation of the NLRC4 inflammasome. Infect. Immun. 79, 1606–1614 (2011).
Franchi, L. Role of inflammasomes in Salmonella infection. Front Microbiol 2, 8 (2011).
Sansonetti, P.J. et al. Caspase-1 activation of IL-1β and IL-18 are essential for Shigella flexneri–induced inflammation. Immunity 12, 581–590 (2000).
Raupach, B., Peuschel, S.K., Monack, D.M. & Zychlinsky, A. Caspase-1-mediated activation of interleukin-1β (IL-1β) and IL-18 contributes to innate immune defenses against Salmonella enterica serovar Typhimurium infection. Infect. Immun. 74, 4922–4926 (2006).
Lara-Tejero, M. et al. Role of the caspase-1 inflammasome in Salmonella Typhimurium pathogenesis. J. Exp. Med. 203, 1407–1412 (2006).
Broz, P. et al. Redundant roles for inflammasome receptors NLRP3 and NLRC4 in host defense against Salmonella. J. Exp. Med. 207, 1745–1755 (2010).
Fink, S.L. & Cookson, B.T. Pyroptosis and host cell death responses during Salmonella infection. Cell. Microbiol. 9, 2562–2570 (2007).
Miao, E.A. et al. Caspase-1-induced pyroptosis is an innate immune effector mechanism against intracellular bacteria. Nat. Immunol. 11, 1136–1142 (2010).
Sauer, J.D. et al. Listeria monocytogenes engineered to activate the NLRC4 inflammasome are severely attenuated and are poor inducers of protective immunity. Proc. Natl. Acad. Sci. USA 108, 12419–12424 (2011).
Warren, S.E. et al. Generation of a Listeria vaccine strain by enhanced caspase-1 activation. Eur. J. Immunol. 41, 1934–1940 (2011).
Akhter, A. et al. Caspase-7 activation by the NLRC4/Ipaf inflammasome restricts Legionella pneumophila infection. PLoS Pathog. 5, e1000361 (2009).
Molofsky, A.B. et al. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J. Exp. Med. 203, 1093–1104 (2006).
Lamkanfi, M. et al. The Nod-like receptor family member Naip5/Birc1e restricts Legionella pneumophila growth independently of caspase-1 activation. J. Immunol. 178, 8022–8027 (2007).
Archer, K.A., Ader, F., Kobayashi, K.S., Flavell, R.A. & Roy, C.R. Cooperation between multiple microbial pattern recognition systems is important for host protection against the intracellular pathogen Legionella pneumophila. Infect. Immun. 78, 2477–2487 (2010).
Martinon, F., Burns, K. & Tschopp, J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol. Cell 10, 417–426 (2002).
Faustin, B. et al. Reconstituted NALP1 inflammasome reveals two-step mechanism of caspase-1 activation. Mol. Cell 25, 713–724 (2007).
Hsu, L.C. et al. A NOD2-NALP1 complex mediates caspase-1-dependent IL-1β secretion in response to Bacillus anthracis infection and muramyl dipeptide. Proc. Natl. Acad. Sci. USA 105, 7803–7808 (2008).
D'Osualdo, A. et al. CARD8 and NLRP1 undergo autoproteolytic processing through a ZU5-like domain. PLoS ONE 6, e27396 (2011).
Boyden, E.D. & Dietrich, W.F. Nalp1b controls mouse macrophage susceptibility to anthrax lethal toxin. Nat. Genet. 38, 240–244 (2006).
Broz, P., von Moltke, J., Jones, J.W., Vance, R.E. & Monack, D.M. Differential requirement for caspase-1 autoproteolysis in pathogen-induced cell death and cytokine processing. Cell Host Microbe 8, 471–483 (2010).
Terra, J.K. et al. Cutting edge: resistance to Bacillus anthracis infection mediated by a lethal toxin sensitive allele of Nalp1b/Nlrp1b. J. Immunol. 184, 17–20 (2010).
Martinon, F., Agostini, L., Meylan, E. & Tschopp, J. Identification of bacterial muramyl dipeptide as activator of the NALP3/cryopyrin inflammasome. Curr. Biol. 14, 1929–1934 (2004).
Marina-García, N. et al. Pannexin-1-mediated intracellular delivery of muramyl dipeptide induces caspase-1 activation via cryopyrin/NLRP3 independently of Nod2. J. Immunol. 180, 4050–4057 (2008).
Kanneganti, T.D. et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature 440, 233–236 (2006).
Bauernfeind, F.G. et al. Cutting edge: NF-κB activating pattern recognition and cytokine receptors license NLRP3 inflammasome activation by regulating NLRP3 expression. J. Immunol. 183, 787–791 (2009).
Harder, J. et al. Activation of the Nlrp3 inflammasome by Streptococcus pyogenes requires streptolysin O and NF-κB activation but proceeds independently of TLR signaling and P2X7 receptor. J. Immunol. 183, 5823–5829 (2009).
Franchi, L., Eigenbrod, T. & Nunez, G. Cutting edge: TNF-α mediates sensitization to ATP and silica via the NLRP3 inflammasome in the absence of microbial stimulation. J. Immunol. 183, 792–796 (2009).
Bauernfeind, F. et al. Cutting edge: reactive oxygen species inhibitors block priming, but not activation, of the NLRP3 inflammasome. J. Immunol. 187, 613–617 (2011).
Embry, C.A., Franchi, L., Nunez, G. & Mitchell, T.C. Mechanism of impaired NLRP3 inflammasome priming by monophosphoryl lipid A. Sci. Signal. 4, ra28 (2011).
Sander, L.E. et al. Detection of prokaryotic mRNA signifies microbial viability and promotes immunity. Nature 474, 385–389 (2011).
Ferrari, D., Chiozzi, P., Falzoni, S., Hanau, S. & Di Virgilio, F. Purinergic modulation of interleukin-1β release from microglial cells stimulated with bacterial endotoxin. J. Exp. Med. 185, 579–582 (1997).
Netea, M.G. et al. Differential requirement for the activation of the inflammasome for processing and release of IL-1β in monocytes and macrophages. Blood 113, 2324–2335 (2009).
Piccini, A. et al. ATP is released by monocytes stimulated with pathogen-sensing receptor ligands and induces IL-1β and IL-18 secretion in an autocrine way. Proc. Natl. Acad. Sci. USA 105, 8067–8072 (2008).
Saitoh, T. et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1β production. Nature 456, 264–268 (2008).
Harris, J. et al. Autophagy controls IL-1β secretion by targeting pro-IL-1β for degradation. J. Biol. Chem. 286, 9587–9597 (2011).
Crisan, T.O. et al. Inflammasome-independent modulation of cytokine response by autophagy in human cells. PLoS ONE 6, e18666 (2011).
Franchi, L., Kanneganti, T.D., Dubyak, G.R. & Nunez, G. Differential requirement of P2X7 receptor and intracellular K+ for caspase-1 activation induced by intracellular and extracellular bacteria. J. Biol. Chem. 282, 18810–18818 (2007).
Petrilli, V. et al. Activation of the NALP3 inflammasome is triggered by low intracellular potassium concentration. Cell Death Differ. 14, 1583–1589 (2007).
Perregaux, D.G. & Gabel, C.A. Human monocyte stimulus-coupled IL-1β posttranslational processing: modulation via monovalent cations. Am. J. Physiol. 275, C1538–C1547 (1998).
Bauernfeind, F. et al. Inflammasomes: current understanding and open questions. Cell Mol. Life Sci. 68, 765–783 (2011).
Muñoz-Planillo, R., Franchi, L., Miller, L.S. & Nunez, G. A critical role for hemolysins and bacterial lipoproteins in Staphylococcus aureus–induced activation of the Nlrp3 inflammasome. J. Immunol. 183, 3942–3948 (2009).
Pelegrin, P. & Surprenant, A. Pannexin-1 mediates large pore formation and interleukin-1β release by the ATP-gated P2X7 receptor. EMBO J. 25, 5071–5082 (2006).
Qu, Y. et al. Pannexin-1 is required for ATP release during apoptosis but not for inflammasome activation. J. Immunol. 186, 6553–6561 (2011).
Duncan, J.A. et al. Neisseria gonorrhoeae activates the proteinase cathepsin B to mediate the signaling activities of the NLRP3 and ASC-containing inflammasome. J. Immunol. 182, 6460–6469 (2009).
Chu, J. et al. Cholesterol-dependent cytolysins induce rapid release of mature IL-1β from murine macrophages in a NLRP3 inflammasome and cathepsin B–dependent manner. J. Leukoc. Biol. 86, 1227–1238 (2009).
Dostert, C. et al. Malarial hemozoin is a Nalp3 inflammasome activating danger signal. PLoS ONE 4, e6510 (2009).
Dostert, C. et al. Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science 320, 674–677 (2008).
Zhou, R., Tardivel, A., Thorens, B., Choi, I. & Tschopp, J. Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat. Immunol. 11, 136–140 (2010).
Masters, S.L. et al. Activation of the NLRP3 inflammasome by islet amyloid polypeptide provides a mechanism for enhanced IL-1β in type 2 diabetes. Nat. Immunol. 11, 897–904 (2010).
Zhou, R., Yazdi, A.S., Menu, P. & Tschopp, J. A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225 (2011).
Nakahira, K. et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 12, 222–230 (2011).
Miller, L.S. et al. MyD88 mediates neutrophil recruitment initiated by IL-1R but not TLR2 activation in immunity against Staphylococcus aureus. Immunity 24, 79–91 (2006).
Miller, L.S. et al. Inflammasome-mediated production of IL-1β is required for neutrophil recruitment against Staphylococcus aureus in vivo. J. Immunol. 179, 6933–6942 (2007).
Toma, C. et al. Pathogenic Vibrio activate NLRP3 inflammasome via cytotoxins and TLR/nucleotide-binding oligomerization domain-mediated NF-κB signaling. J. Immunol. 184, 5287–5297 (2010).
Witzenrath, M. et al. The NLRP3 inflammasome is differentially activated by pneumolysin variants and contributes to host defense in pneumococcal pneumonia. J. Immunol. 187, 434–440 (2011).
McNeela, E.A. et al. Pneumolysin activates the NLRP3 inflammasome and promotes proinflammatory cytokines independently of TLR4. PLoS Pathog. 6, e1001191 (2010).
Hoegen, T. et al. The NLRP3 inflammasome contributes to brain injury in pneumococcal meningitis and is activated through ATP-dependent lysosomal cathepsin B release. J. Immunol. 187, 5440–5451 (2011).
Kayagaki, N. et al. Non-canonical inflammasome activation targets caspase-11. Nature 479, 117–121 (2011).
Ng, J. et al. Clostridium difficile toxin-induced inflammation and intestinal injury are mediated by the inflammasome. Gastroenterology 139, 542–552 (2010).
Brodsky, I.E. et al. A Yersinia effector protein promotes virulence by preventing inflammasome recognition of the type III secretion system. Cell Host Microbe 7, 376–387 (2010).
Zheng, Y. et al. A Yersinia effector with enhanced inhibitory activity on the NF-κB pathway activates the NLRP3/ASC/caspase-1 inflammasome in macrophages. PLoS Pathog. 7, e1002026 (2011).
Kanneganti, T.D. et al. Critical role for cryopyrin/Nalp3 in activation of caspase-1 in response to viral infection and double-stranded RNA. J. Biol. Chem. 281, 36560–36568 (2006).
Muruve, D.A. et al. The inflammasome recognizes cytosolic microbial and host DNA and triggers an innate immune response. Nature 452, 103–107 (2008).
Ichinohe, T., Pang, I.K. & Iwasaki, A. Influenza virus activates inflammasomes via its intracellular M2 ion channel. Nat. Immunol. 11, 404–410 (2010).
Ichinohe, T., Lee, H.K., Ogura, Y., Flavell, R. & Iwasaki, A. Inflammasome recognition of influenza virus is essential for adaptive immune responses. J. Exp. Med. 206, 79–87 (2009).
Thomas, P.G. et al. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity 30, 566–575 (2009).
Allen, I.C. et al. The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity 30, 556–565 (2009).
Gross, O. et al. Syk kinase signalling couples to the Nlrp3 inflammasome for anti-fungal host defence. Nature 459, 433–436 (2009).
Joly, S. et al. Cutting edge: Candida albicans hyphae formation triggers activation of the Nlrp3 inflammasome. J. Immunol. 183, 3578–3581 (2009).
Hise, A.G. et al. An essential role for the NLRP3 inflammasome in host defense against the human fungal pathogen Candida albicans. Cell Host Microbe 5, 487–497 (2009).
Tomalka, J. et al. A novel role for the NLRC4 inflammasome in mucosal defenses against the fungal pathogen Candida albicans. PLoS Pathog. 7, e1002379 (2011).
Shio, M.T. et al. Malarial hemozoin activates the NLRP3 inflammasome through Lyn and Syk kinases. PLoS Pathog. 5, e1000559 (2009).
Reimer, T. et al. Experimental cerebral malaria progresses independently of the Nlrp3 inflammasome. Eur. J. Immunol. 40, 764–769 (2010).
Franchi, L., Munoz-Planillo, R., Reimer, T., Eigenbrod, T. & Nunez, G. Inflammasomes as microbial sensors. Eur. J. Immunol. 40, 611–615 (2010).
Rathinam, V.A. et al. The AIM2 inflammasome is essential for host defense against cytosolic bacteria and DNA viruses. Nat. Immunol. 11, 395–402 (2010).
Meixenberger, K. et al. Listeria monocytogenes–infected human peripheral blood mononuclear cells produce IL-1β, depending on listeriolysin O and NLRP3. J. Immunol. 184, 922–930 (2010).
Kim, S. et al. Listeria monocytogenes is sensed by the NLRP3 and AIM2 inflammasome. Eur. J. Immunol. 40, 1545–1551 (2010).
Hara, H. et al. Dependency of caspase-1 activation induced in macrophages by Listeria monocytogenes on cytolysin, listeriolysin O, after evasion from phagosome into the cytoplasm. J. Immunol. 180, 7859–7868 (2008).
Ozören, N. et al. Distinct roles of TLR2 and the adaptor ASC in IL-1β/IL-18 secretion in response to Listeria monocytogenes. J. Immunol. 176, 4337–4342 (2006).
Tsuji, N.M. et al. Roles of caspase-1 in Listeria infection in mice. Int. Immunol. 16, 335–343 (2004).
Willingham, S.B. et al. Microbial pathogen-induced necrotic cell death mediated by the inflammasome components CIAS1/cryopyrin/NLRP3 and ASC. Cell Host Microbe 2, 147–159 (2007).
Smith, P.D. et al. Intestinal macrophages and response to microbial encroachment. Mucosal Immunol. 4, 31–42 (2011).
Willingham, S.B. et al. NLRP3 (NALP3, cryopyrin) facilitates in vivo caspase-1 activation, necrosis, and HMGB1 release via inflammasome-dependent and -independent pathways. J. Immunol. 183, 2008–2015 (2009).
Dunne, A. et al. Inflammasome activation by adenylate cyclase toxin directs Th17 responses and protection against Bordetella pertussis. J. Immunol. 185, 1711–1719 (2010).
Brereton, C.F. et al. Escherichia coli heat-labile enterotoxin promotes protective Th17 responses against infection by driving innate IL-1 and IL-23 production. J. Immunol. 186, 5896–5906 (2011).
Shimada, K. et al. Caspase-1 dependent IL-1β secretion is critical for host defense in a mouse model of Chlamydia pneumoniae lung infection. PLoS ONE 6, e21477 (2011).
He, X. et al. Inflammation and fibrosis during Chlamydia pneumoniae infection is regulated by IL-1 and the NLRP3/ASC inflammasome. J. Immunol. 184, 5743–5754 (2010).
Master, S.S. et al. Mycobacterium tuberculosis prevents inflammasome activation. Cell Host Microbe 3, 224–232 (2008).
Koo, I.C. et al. ESX-1-dependent cytolysis in lysosome secretion and inflammasome activation during mycobacterial infection. Cell Microbiol. 10, 1866–1878 (2008).
Mishra, B.B. et al. Mycobacterium tuberculosis protein ESAT-6 is a potent activator of the NLRP3/ASC inflammasome. Cell Microbiol. 12, 1046–1063 (2010).
Mariathasan, S. et al. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 440, 228–232 (2006).
Warren, S.E., Mao, D.P., Rodriguez, A.E., Miao, E.A. & Aderem, A. Multiple Nod-like receptors activate caspase 1 during Listeria monocytogenes infection. J. Immunol. 180, 7558–7564 (2008).
Wu, J., Fernandes-Alnemri, T. & Alnemri, E.S. Involvement of the AIM2, NLRC4, and NLRP3 inflammasomes in caspase-1 activation by Listeria monocytogenes. J. Clin. Immunol. 30, 693–702 (2010).
Tsuchiya, K. et al. Involvement of absent in melanoma 2 in inflammasome activation in macrophages infected with Listeria monocytogenes. J. Immunol. 185, 1186–1195 (2010).
Gurcel, L., Abrami, L., Girardin, S., Tschopp, J. & van der Goot, F.G. Caspase-1 activation of lipid metabolic pathways in response to bacterial pore-forming toxins promotes cell survival. Cell 126, 1135–1145 (2006).
Galle, M. et al. The Pseudomonas aeruginosa type III secretion system plays a dual role in the regulation of caspase-1 mediated IL-1β maturation. J. Cell Mol. Med. 12, 1767–1776 (2007).
Lightfield, K.L. et al. Differential requirements for NAIP5 in activation of the NLRC4 (IPAF) inflammasome. Infect. Immun. 79, 1606–1614 (2011).
Kofoed, E.M. & Vance, R.E. Innate immune recognition of bacterial ligands by NAIPs determines inflammasome specificity. Nature 477, 592–595 (2011).
Nour, A.M. et al. Anthrax lethal toxin triggers the formation of a membrane-associated inflammasome complex in murine macrophages. Infect. Immun. 77, 1262–1271 (2009).
Fernandes-Alnemri, T. et al. The AIM2 inflammasome is critical for innate immunity to Francisella tularensis. Nat. Immunol. 11, 385–393 (2010).
Saïd-Sadier, N., Padilla, E., Langsley, G. & Ojcius, D.M. Aspergillus fumigatus stimulates the NLRP3 inflammasome through a pathway requiring ROS production and the Syk tyrosine kinase. PLoS ONE 5, e10008 (2010).
Nour, A.M. et al. Varicella-zoster virus infection triggers formation of an interleukin-1β (IL-1β)-processing inflammasome complex. J. Biol. Chem. 286, 17921–17933 (2011).
Hornung, V. et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 458, 514–518 (2009).
Acknowledgements
We thank G. Chen for critically reading the manuscript. Supported by the US National Institutes of Health (R01 AI0647748, R01 Al063331 and R01 DK61707) and the Crohn's and Colitis Foundation of America (L.F.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Franchi, L., Muñoz-Planillo, R. & Núñez, G. Sensing and reacting to microbes through the inflammasomes. Nat Immunol 13, 325–332 (2012). https://doi.org/10.1038/ni.2231
Published:
Issue Date:
DOI: https://doi.org/10.1038/ni.2231
This article is cited by
-
Whole transcriptome sequencing for revealing the pathogenesis of sporotrichosis caused by Sporothrix globosa
Scientific Reports (2024)
-
Control of complement-induced inflammatory responses to SARS-CoV-2 infection by anti-SARS-CoV-2 antibodies
The EMBO Journal (2024)
-
Involvement of NLRP3 inflammasome pathway in the protective mechanisms of ferulic acid and p-coumaric acid in LPS-induced sickness behavior and neuroinflammation in mice
Naunyn-Schmiedeberg's Archives of Pharmacology (2024)
-
Lactobacilli Probiotics Modulate Antibacterial Response Gene Transcription of Dendritic Cells Challenged with LPS
Probiotics and Antimicrobial Proteins (2024)
-
VCP Inhibition Augments NLRP3 Inflammasome Activation
Inflammation (2024)